Applications
If you are a synthetic biologist, you may not be aware that there are already many software tools available that support different aspects of the synthetic biology workflow, such as optimizing DNA assembly or simulating gene networks. Each of the software tools listed here supports exchange of genetic designs in the SBOL file format (SBOL core) or supports display of genetic designs using standard SBOL Visual glyphs.
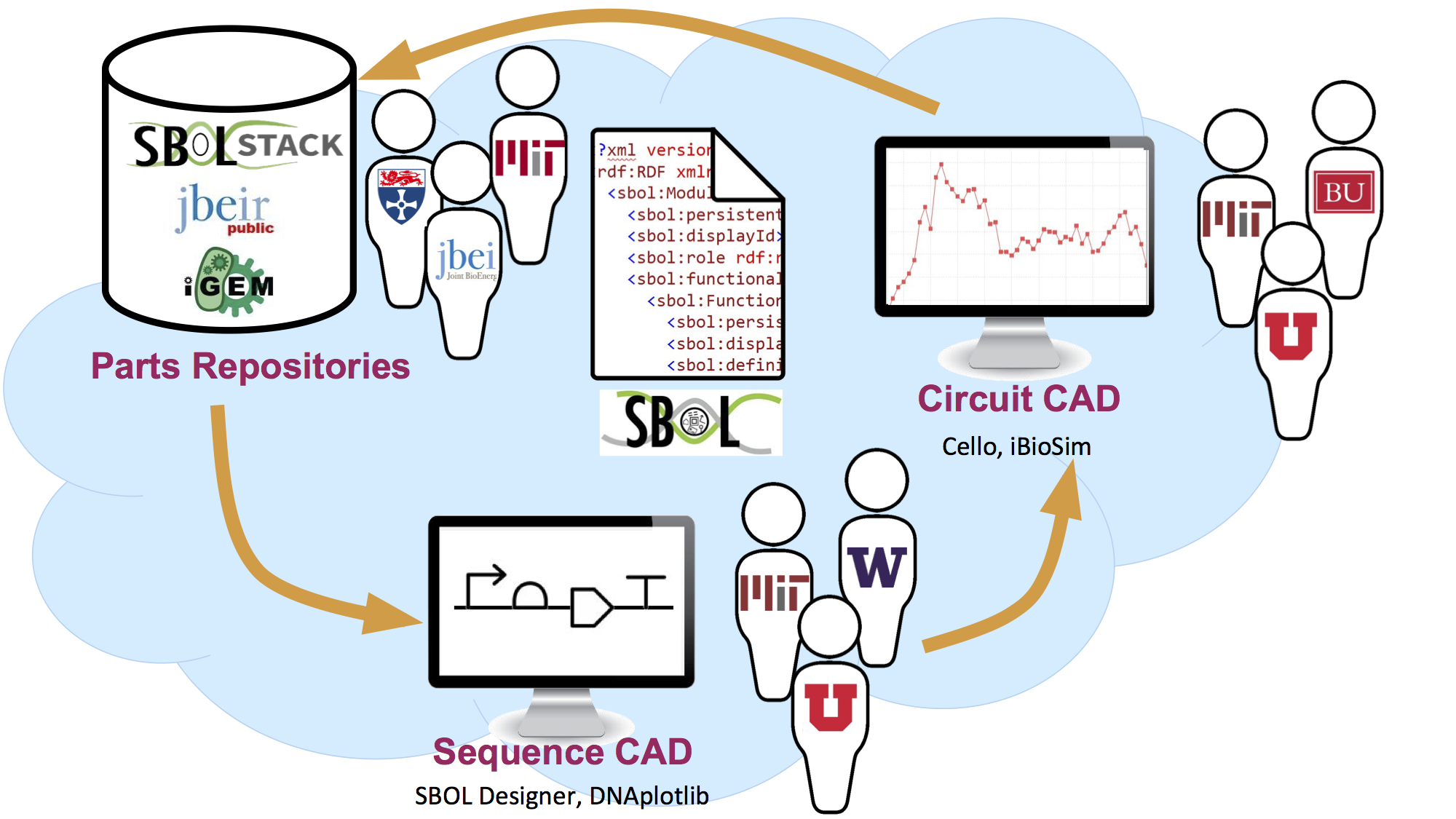
If you would like us to feature your application here, please fill out this survey.
GenoCAD
A rule-based DNA design tool.
Graphviz
a package of open-source tools initiated by AT&T Labs Research for drawing graphs specified in DOT language scripts.
iBioSim
iBioSim has been developed for the modeling, analysis, and design of genetic circuits.
ICE
Physical and Informatic repository and associated tools for plant seeds, microbial strains, DNA and protein sequences.
j5
DNA assembly design automation
MoSeC
Automated derivation of genetic circuit designs from computational models.
Parts & Pools
A graphical, drag and drop design of synthetic gene circuit with Standard Biological Parts.
Pigeon Visualizer
A tool for creating SBOL Visual diagrams from a concise textual representation.
Pinecone
Serotiny’s web-based protein design software aimed at researchers doing R&D for therapeutics, materials, as well as those in basic research.
Pool Designer
Designing Multiplexed Pools for Assembling Library Constructs