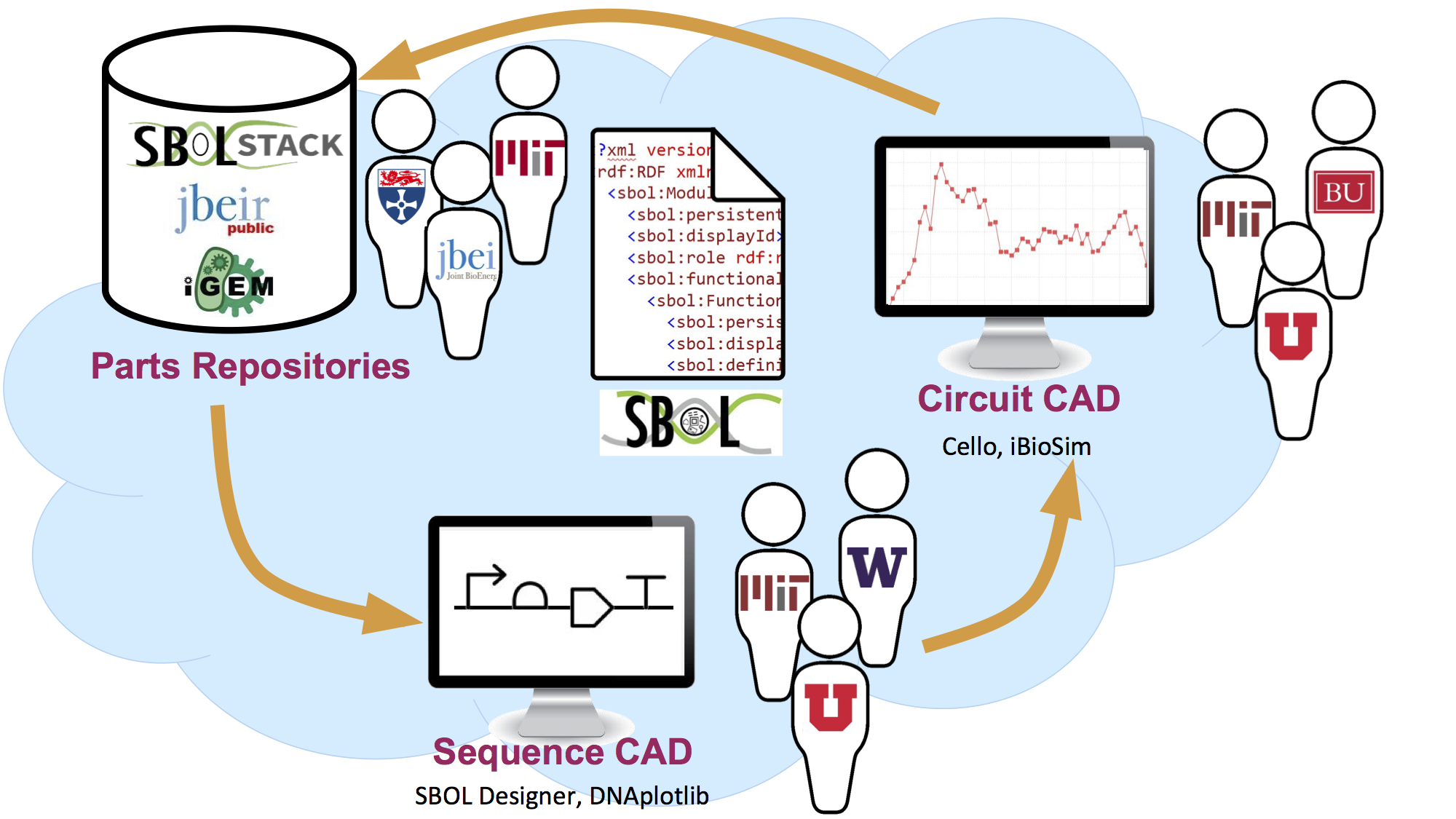
Applications
If you are a synthetic biologist, you may not be aware that there are already many software tools available that support different aspects of the synthetic biology workflow, such as optimizing DNA assembly or simulating gene networks. Each of the software tools listed here supports exchange of genetic designs in the SBOL file format (SBOL core) or supports display of genetic designs using standard SBOL Visual glyphs.
If you would like us to feature your application here, please fill out this survey.
SBOLCanvas
A web application for creation and editing of genetic constructs using the SBOL data and visual standard.
BOOST
A platform of tools to design DNA sequences for manufacturing
Cello
A tool for Genetic circuit design automation
Device Editor
Visual biocad canvas for specifying combinatorial DNA constructs for fabrication
DNAplotlib
DNAplotlib enables highly customizable visualization of individual genetic constructs and libraries of design variants. It can be thought of in many ways as matplotlib for genetic diagrams.
Eugene
A textual specification language for the rule-based design of synthetic biological systems, devices, parts, and DNA sequences.
Finch
Rule-based generation of constructs
Flapjack
Flapjack is a data management and analysis app for genetic circuit characterization to store, share, mix, analyze and plot your SynBio data
GeneGenie
GeneGenie supports the design and self-assembly of synthetic genes and constructs.
GeneTech
A tool which allows a user to generate genetic logic circuits only by specifying the logical function desired to be achieved in a living cell.