SBOL Examples
2019 (Data Model Examples)
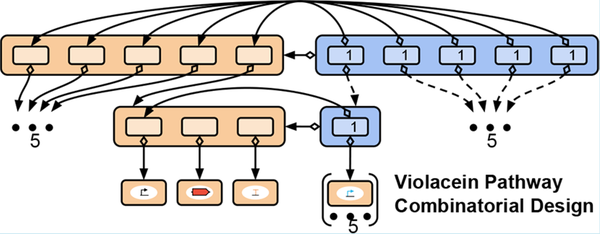
Specifying Combinatorial Designs with the Synthetic Biology Open Language (SBOL)
As improvements in DNA synthesis technology and assembly methods make combinatorial assembly of genetic constructs increasingly accessible, methods for representing genetic constructs likewise need to improve to handle the exponential growth of combinatorial design space. To this end, we present a community accepted extension of the SBOL data standard that allows for the efficient and flexible encoding of combinatorial designs. This extension includes data structures for representing genetic designs with “variable” components that can be implemented by choosing one of many linked designs for existing genetic parts or constructs. We demonstrate the representational power of the SBOL combinatorial design extension through case studies on metabolic pathway design and genetic circuit design, and we report the expansion of the SBOLDesigner software tool to support users in creating and modifying combinatorial designs in SBOL.
SBOL 2.0 Examples
CRISPR Repression Model SBOL file example
Genetic Toggle Switch example
Four-input AND gate example
SBOL 2.0 Specification examples
GeneticToggleSwitch Systems Biology Model example
Directed Acyclic Genetic Circuit examples
Further SBOL 2.0 examples are located here. This comprehensive set of data files includes in addition to more SBOL 2.0 files, SBOL 1.1 files used to test conversion to SBOL 2.0, GenBank files used to test conversion to SBOL 2.0, and invalid SBOL 2.0 files used to test validation.
SBOL 1.1 Examples
NBT Examples: pIKE and pTAK_cassettes
NBT Example: pIKE and pTAK partial cassettes
NBT Example: pIKE and pTAK toggle switches