SBOL Visual Examples
2020 (Visual Examples)
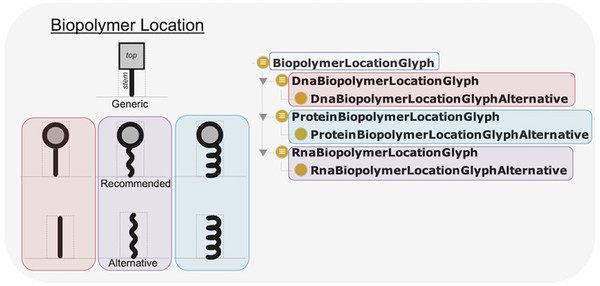
SBOL Visual 2 Ontology
Standardizing the visual representation of genetic parts and circuits is essential for unambiguously creating and interpreting genetic designs. To this end, an increasing number of tools are adopting well-defined glyphs from the Synthetic Biology Open Language (SBOL) Visual standard to represent various genetic parts and their relationships. However, the implementation and maintenance of the relationships between biological elements or concepts and their associated glyphs has up to now been left up to tool developers. We address this need with the SBOL Visual 2 Ontology, a machine-accessible resource that provides rules for mapping from genetic parts, molecules, and interactions between them, to agreed SBOL Visual glyphs. This resource, together with a web service, can be used as a library to simplify the development of visualization tools, as a stand-alone resource to computationally search for suitable glyphs, and to help facilitate integration with existing biological ontologies and standards in synthetic biology.
Synthetic biology open language visual (SBOL visual) version 2.2
People who are engineering biological organisms often find it useful to communicate in diagrams, both about the structure of the nucleic acid sequences that they are engineering and about the functional relationships between sequence features and other molecular species. Some typical practices and conventions have begun to emerge for such diagrams. The Synthetic Biology Open Language Visual (SBOL Visual) has been developed as a standard for organizing and systematizing such conventions in order to produce a coherent language for expressing the structure and function of genetic designs. This document details version 2.2 of SBOL Visual, which builds on the prior SBOL Visual 2.1 in several ways. First, the grounding of molecular species glyphs is changed from BioPAX to SBO, aligning with the use of SBO terms for interaction glyphs. Second, new glyphs are added for proteins, introns, and polypeptide regions (e. g., protein domains), the prior recommended macromolecule glyph is deprecated in favor of its alternative, and small polygons are introduced as alternative glyphs for simple chemicals.
2019 (Visual Examples)
2015 (Visual Examples)
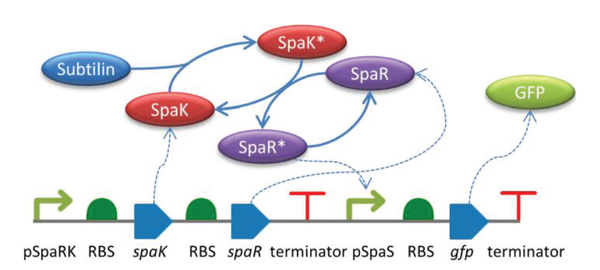
Composable Modular Models for Synthetic Biology
Modelling and computational simulation are crucial for the large-scale engineering of biological circuits since they allow the system under design to be simulated prior to implementation in vivo. To support automated, model-driven design it is desirable that in silico models are modular, composable and use standard formats. The synthetic biology design process typically involves the composition of genetic circuits from individual parts. At the most basic level, these parts are representations of genetic features such as promoters, ribosome binding sites (RBSs), and coding sequences (CDSs). However, it is also desirable to model the biological molecules and behaviour that arise when these parts are combined in vivo. Modular models of parts can be composed and their associated systems simulated, facilitating the process of model-centred design. The availability of databases of modular models is essential to support software tools used in the model-driven design process. In this article, we present an approach to support the development of composable, modular models for synthetic biology, termed Standard Virtual Parts. We then describe a programmatically accessible and publicly available database of these models to allow their use by computational design tools.
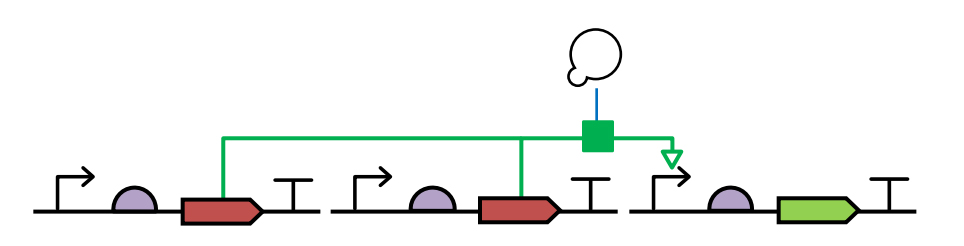
Examples to demonstrate the usage of SBOL Visual glyphs is given below.
Example1: A SBOL visual design of the circuit that contains a promoter, RBS, CDS, and terminator. The promoter is stimulated by CDS that it regulates.
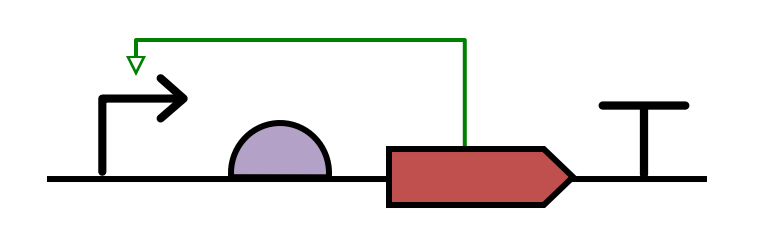
Example2: Phosphorylation of an inactive transcription factor (produced by two different CDSs) by a kinase to form an active transcriptional activator, which then stimulates a promoter.